Section: New Results
Computational Physiology
Biophysical Modeling and Simulation of Longitudinal Brain MRIs with Atrophy in Alzheimer's Disease
Participants : Bishesh Khanal [correspondent] , Nicholas Ayache, Xavier Pennec.
This work has been partly supported by the European Research Council through the ERC Advanced Grant MedYMA (on Biophysical Modeling and Analysis of Dynamic Medical Images).
Alzheimer's Disease (AD), modeling brain deformation, biophysical model, simulation
-
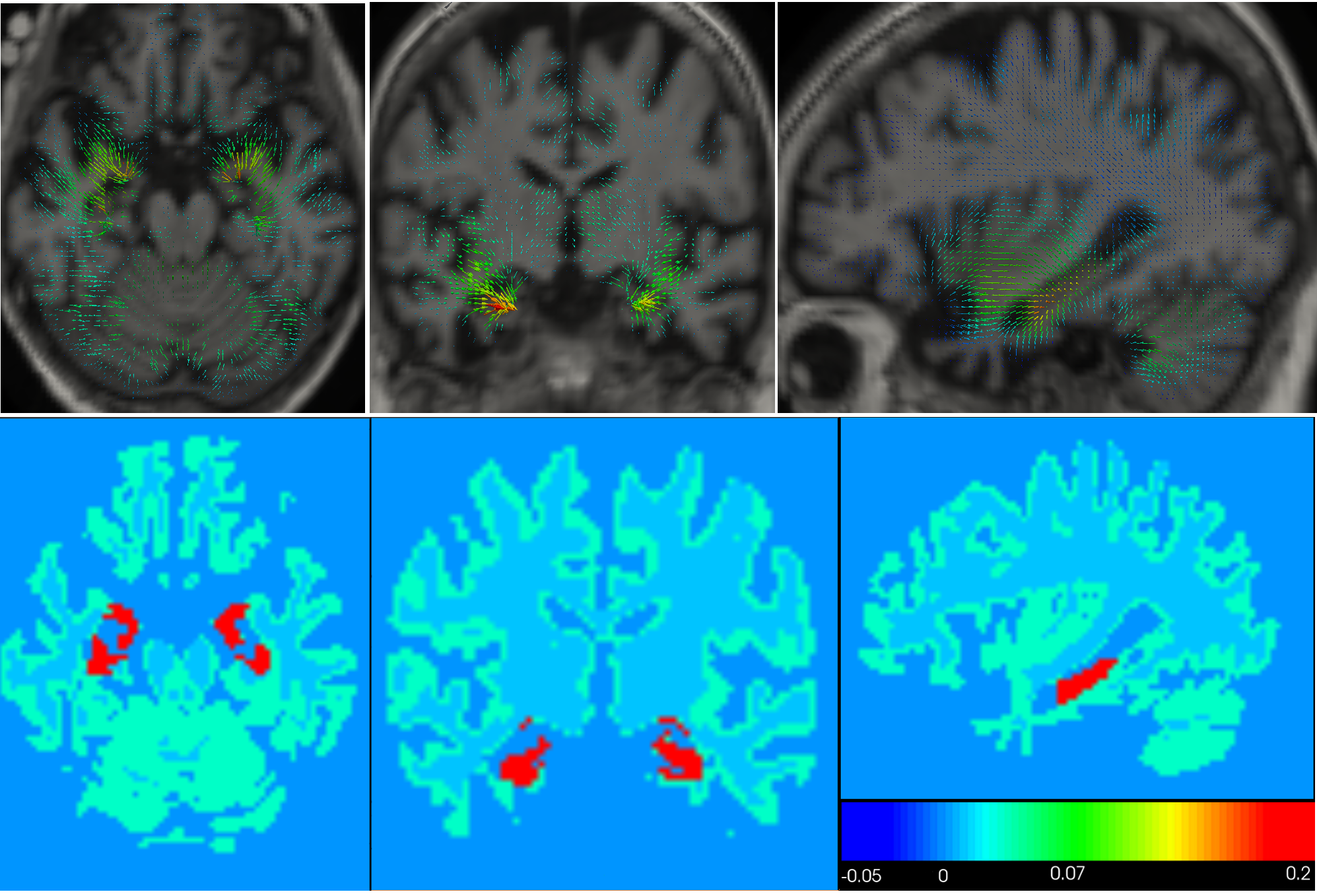
We propose a biophysical model of brain deformation due to atrophy in Alzheimer's Disease(AD) [17] . The model allows simulation of longitudinal brain MRIs with a desired level of atrophy in brain parenchyma. Here we enhanced our previous implementation to model brain parenchyma and cerebrospinal fluid (CSF) differently so that there is no need to prescribe atrophy in CSF region (see Fig.13 ).
-
The model could be used to explore different possible hypotheses about evolution of atrophy in the brain and how it affects the brain shape changes.
|
Glioblastoma : Study of the vasogenic edema
Participants : Matthieu Lê [correspondent] , Hervé Delingette, Jan Unkelbach [Massachussetts General Hospital] , Nicholas Ayache.
This work is carried out between Ascelpios research group, Inria Sophia Antipolis, France and the Department of Radiation Oncology of the Massachusetts General Hospital, Boston, USA.
Glioblastoma, Vasogenic Edema, Radiotherapy, Target Delineation
-
We studied the impact of anti-angiogenic treatment on the MRI appearance of glioblastoma.
-
We studied how MRI extracted features could help distinguish between the vasogenic edema and the tumor infiltration[22] .
-
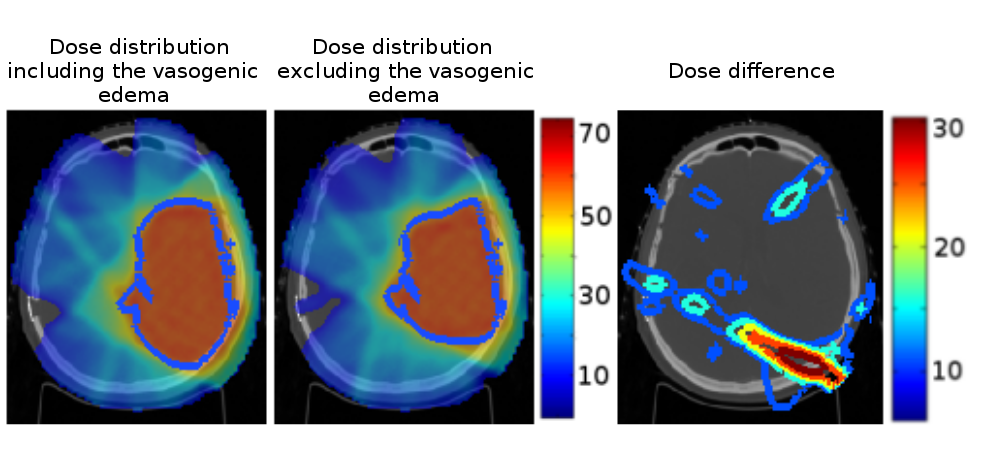
We analyzed the impact of excluding the vasogenic edema from the gross tumor volume during radiation therapy (see Fig. 14 ). Our approach leads to a dose more comformal to the underlying tumor cell density knowing that prescribing less dose might open the way for later re-irradiation.
|
Image-based Prediction of Cardiac Ablation Targets
Participants : Rocio Cabrera Lozoya [correspondent] , Maxime Sermesant, Nicholas Ayache.
Electrophysiology, ablation planning, machine learning
Ventricular radio-frequency ablation (RFA) can have a critical impact on preventing sudden cardiac arrest but is challenging due to a highly complex arrhythmogenic substrate. We aim at identifying local image characteristics capable of predicting the presence of local abnormal ventricular activities (LAVA). This could lead to pre-operatively and non-invasively improve and accelerate the procedure.
-
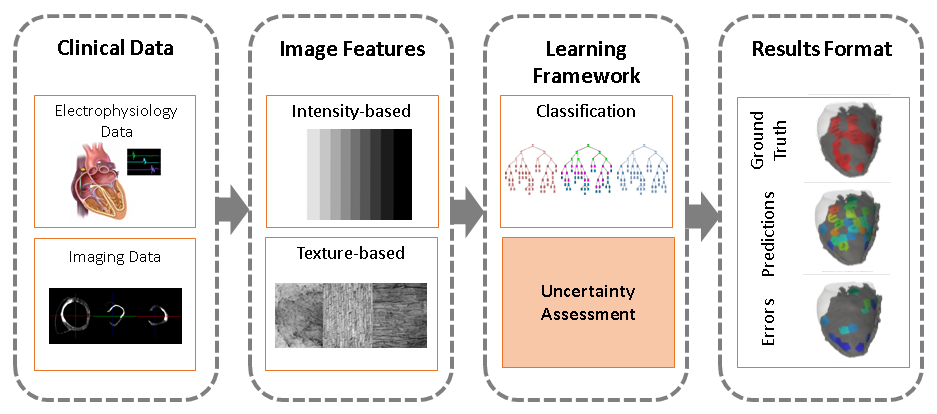
We present the use of intensity and texture-based local imaging features in the vicinity of myocardial scar and grey zones towards the prediction of RFA target localisation (see Fig.15 ).
-
We detail the uncertainty in the data and explore its impact on the classification results.
-
A preliminary output with visual interpretation and potential use in a clinical environment was presented.
-
The encouraging obtained results warrant further investigation and open up possibilities for non-invasive cardiac arrhythmia ablation planning. [13]
|
Personalised Canine Electromechanical Model of the Heart
Participants : Sophie Giffard-Roisin [correspondent] , Maxime Sermesant, Hervé Delingette, Stéphanie Marchesseau, Nicholas Ayache.
This work has been supported by the European Project FP7 under grant agreement VP2HF (no 611823) and the ERC Advanced Grant MedYMA (on Biophysical Modeling and Analysis of Dynamic Medical Images).
Cardiac Modelling, Personalised Simulation, Electrical and Mechanical Simulation
-
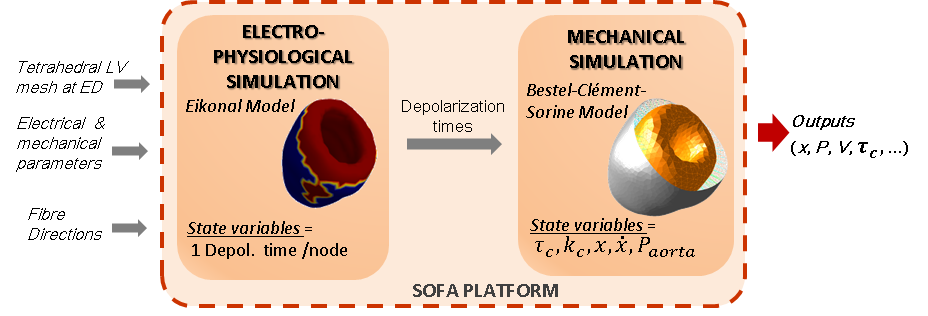
We studied the coupled electro-mechanical modelling of the heart, where the mechanics is handled by the Bestel-Clement-Sorine model while the electrophysiological phenomena is driven by an Eikonal model (see Fig. 16 ).
-
We participated to the STACOM'2014 LV Mechanics Challenge in Boston[15] where four healthy canine clinical data (left ventricles) were provided. The validation was performed on local displacements. Our model has been calibrated by a quantitative sensitivity study as well as a personalized automatic calibration.
|
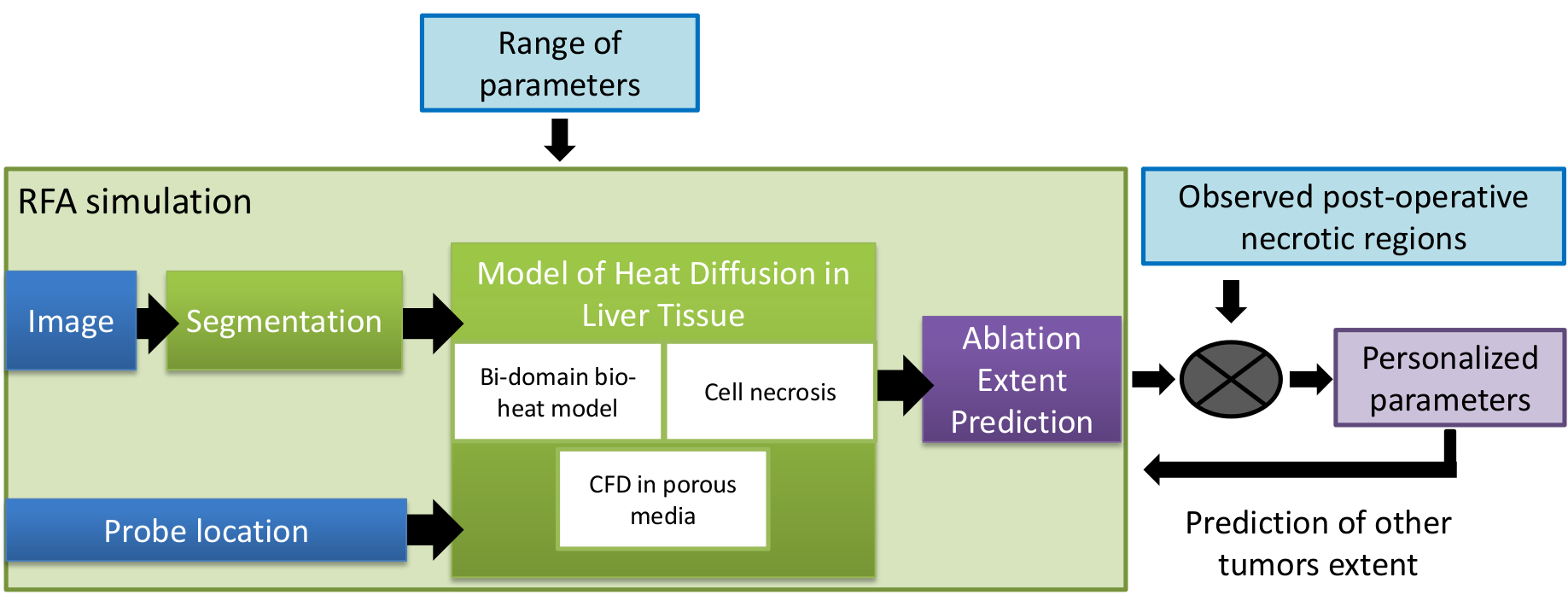
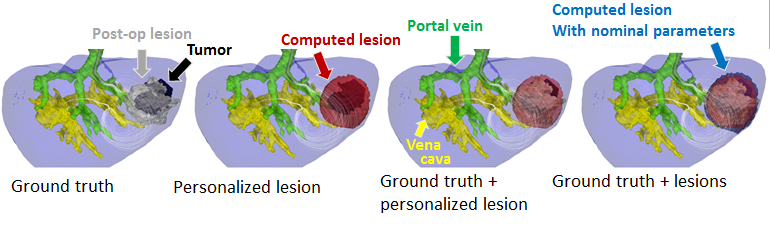
Computational modeling of radiofrequency ablation for the planning and guidance of abdominal tumor treatment
Participants : Chloé Audigier [correspondent] , Hervé Delingette, Tommaso Mansi, Nicholas Ayache.
This PhD is carried out between Asclepios research group, Inria Sophia Antipolis and the Image Analytics and Informatics global field, Siemens Corporate Research, Princeton, USA.
Radio Frequency Abation, Patient-Specific Simulation, Lattice Boltzmann Method, Computational Fluid Dynamics, Heat Transfer, Therapy Planning, Liver
Radiofrequency abation (RFA) is a minimally invasive therapy suited for liver tumor ablation. However a patient-specific predicitive tool to plan and guide the treatment is required. We developed a computational framework for patient-specific planning of RFA (see Fig.17 ) :
-
A patient-specific detailed anatomical model of the liver is estimated from standard CT image and meshed to generate a tetrahedral volume mesh. The structures of interest include the parenchyma, lesion, hepatic vein and vena cava.
-
A Computation Fluic Dynamic and porous media solver using the Lattice Boltzmann Method is used to compute the patient-specific blood flow in the hepatic circulatory system and the blood flow distribution inside the parenchyma.
-
Bio-heat equation has been implemented with a Lattice Boltmann Method also to model efficiently the heat propagation in biological tissues accounting for the cooling effect of neighboring vessels. A cell death model have been combined to account for the cellular necrosis.
Then this forward model is used to estimate patient-specific model parameters as presented in the ABDI workshop at MICCAI 2014 [12] . This work presented obtained the best paper award of the workshop.
|
|
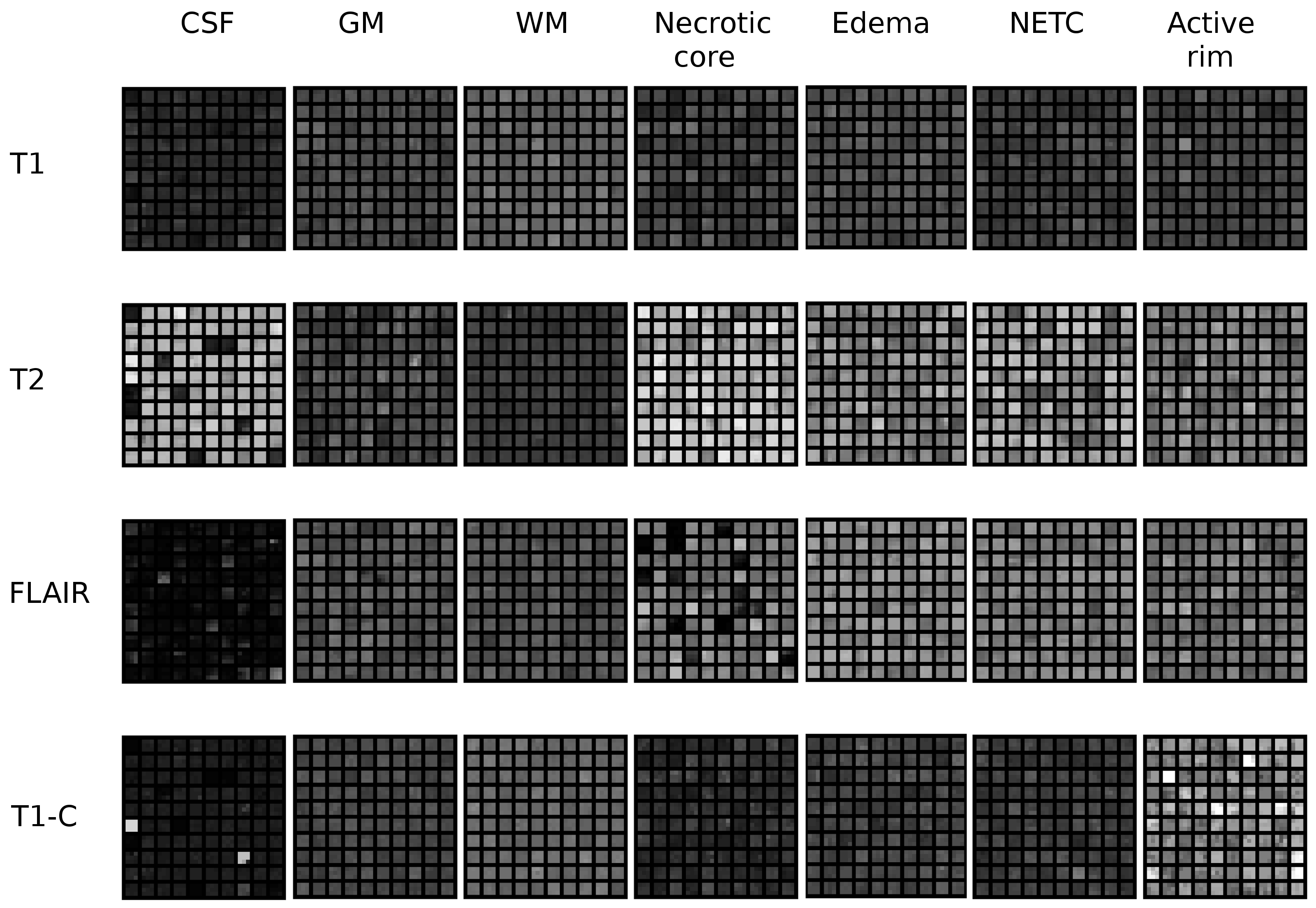
Multi-channel patch-based glioma segmentation
Participants : Nicolas Cordier [correspondent] , Hervé Delingette, Nicholas Ayache.
Part of this work was funded by the European Research Council through the ERC Advanced Grant MedYMA (on Biophysical Modeling and Analysis of Dynamic Medical Images).
Brain, MRI, Glioma, Patch-based Segmentation, Tumor Simulation
The segmentation of glioblastoma, the most severe case of brain tumors, is a crucial step for diagnostic assessment and therapy planning. In order to perform the manual delineation of the tumor compartments, the clinicians have to concurrently screen multi-channel 3D MRI, which makes the process both time-consuming and subject to inter-expert delineation variability. We are building upon the patch-based segmentation framework, the state-of-the-art for the segmentation of healthy brain structures, to present automatic glioma segmentation algorithms. Our 2013 submission to the MICCAI Brain Tumor Segmentation Challenge has been improved by:
-
replacing the heuristic label fusion strategy with a more robust approach,
-
integrating information such as statistics of appearance and position,
-
filtering out the training patches for which the labels are less reliable (see Fig.19 ).
|